library(tidyverse) # process data
library(tidybayes) # get model draws in a tidy format
library(brms) # fit bayesian regression models
library(see) # colour palette
# set the theme
theme_set(theme_bw() +
theme(legend.position = "bottom"))The consequence of ignoring censorship
The title is perhaps misleading, I’m not about to discuss anything about censorship in terms of politics. For better or worse, this is a post about censorship in the statistical sense of the word. So, depending on your view point, this is maybe a more exciting topic? Maybe it doesn’t quite make people feel quite as animated, but then again maybe it should. Censorship in statistics is a genuinely interesting problem to consider, and has some fairly important consequences if ignored or not handled with care.
Library
Before we go anywhere, here is a pretty standard set of packages, it’s not like we’re doing anything insane just now.
Introduction
Missing follow up data is very common in healthcare research. There are many reasons why we might not have complete follow up from people: simple reasons like they only joined the study population recently and so we don’t have extensive records, frustrating reasons like they moved and we no longer have access to their data, and morbid reasons like death. In each of these cases, we’re only able to explain what happened to them for the time we observed them, and this is fine.
However, despite this being a constant presence in the world of healthcare research, it often feels like this goes under appreciated. Despite there being many hard working people engaged in trying to help us gain insight from censored data, there appears to be a lack of willingness to engage with this problem. It is all too common to see people attempting to answer questions that should employ survival techniques using inappropriate methods that ignore the censoring or, perhaps even worse, remove anyone that doesn’t have sufficient follow up.
As the pedant I am, I wanted to make something quick to demonstrate why this is a problem and hopefully show people why they should care about this when running any analysis. This is by no means the most intensive guide to survival analysis, but more something to point to should I be asked to ignore this issue when running my own analysis.
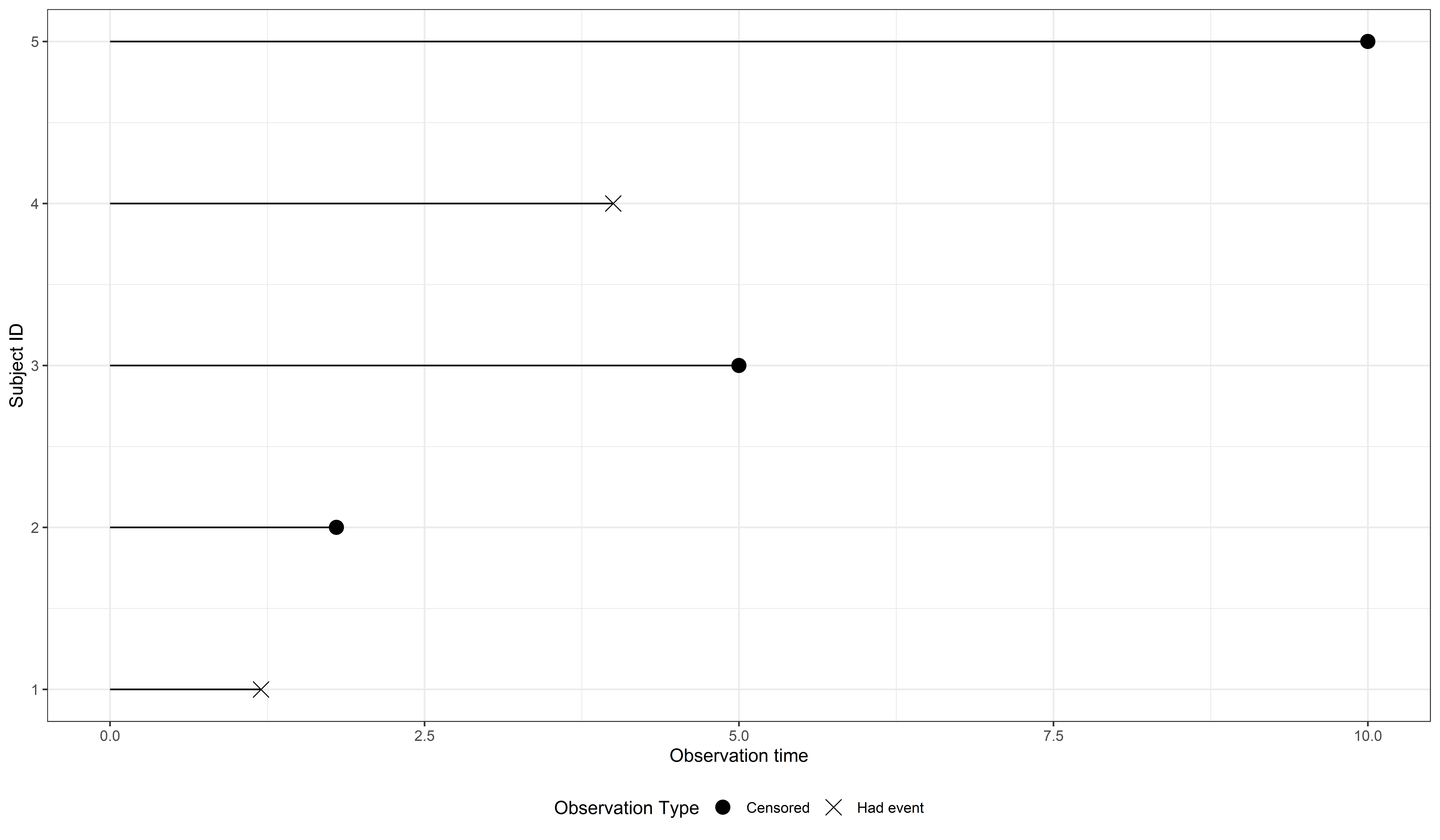
Censorship of data can take several different forms, but the most common is called “right censored”. This is when we “know” the start data and continued to observed each subject until they had the event or time ran out. For those that have the event, we can say when the even occurred. However, for those that are censored all we can say is that they were event free for at least \(X\) amount of time.
Data of this sort could look something like this:
Code
tibble(subj = 1:5,
survTime = c(1.2, 1.8, 5, 4, 10),
obsType = c(0, 1, 1, 0, 1)) %>%
mutate(obsLabel = ifelse(obsType, "Censored", "Had event")) %>%
ggplot(aes(survTime, subj, shape = obsLabel)) +
geom_segment(aes(x = 0, xend = survTime, yend = subj)) +
geom_point(size = 4) +
scale_shape_manual(values = c(16, 4)) +
scale_x_continuous("Observation time") +
scale_y_continuous("Subject ID") +
labs(shape = "Observation Type")In the real world, there are plenty of reasons why someone could be censored; subjects could leave the study, the study could end before observing the event for some subjects, or the subject could experience another event that means they cannot experience the event of interest. The last in this list is referred to as a competing risk and is something to discuss for another time, this point is to simply demonstrate that censorhip has many flavours and should be handled with care.
Making some data
I’m going to use a Weibull distribution as the basis for my synthetic data. This is often used in survival analysis as it has a fully parametric form and can be very useful. There are plenty of others to choose from, but I like this one. You should be able to follow this and simply replace the word “Weibull” with any other distribution that is used in survival analysis.
The simulated dataset will have two groups that we want to compare to each other, with one group being more likely to experience the event earlier. For now, we’ll imagine that we’re looking at people who have a condition versus those that don’t, and their risk of death. We can get into why not applying a survival analysis here would lead to weird conclusions at some point, but for now, we can just make the data.
# set seed for reproducibility
set.seed(87651)
# setup data
N <- 400
# setup parameters for cases and controls
cont_shape <- 2
cont_scale <- 10
case_shape <- .9
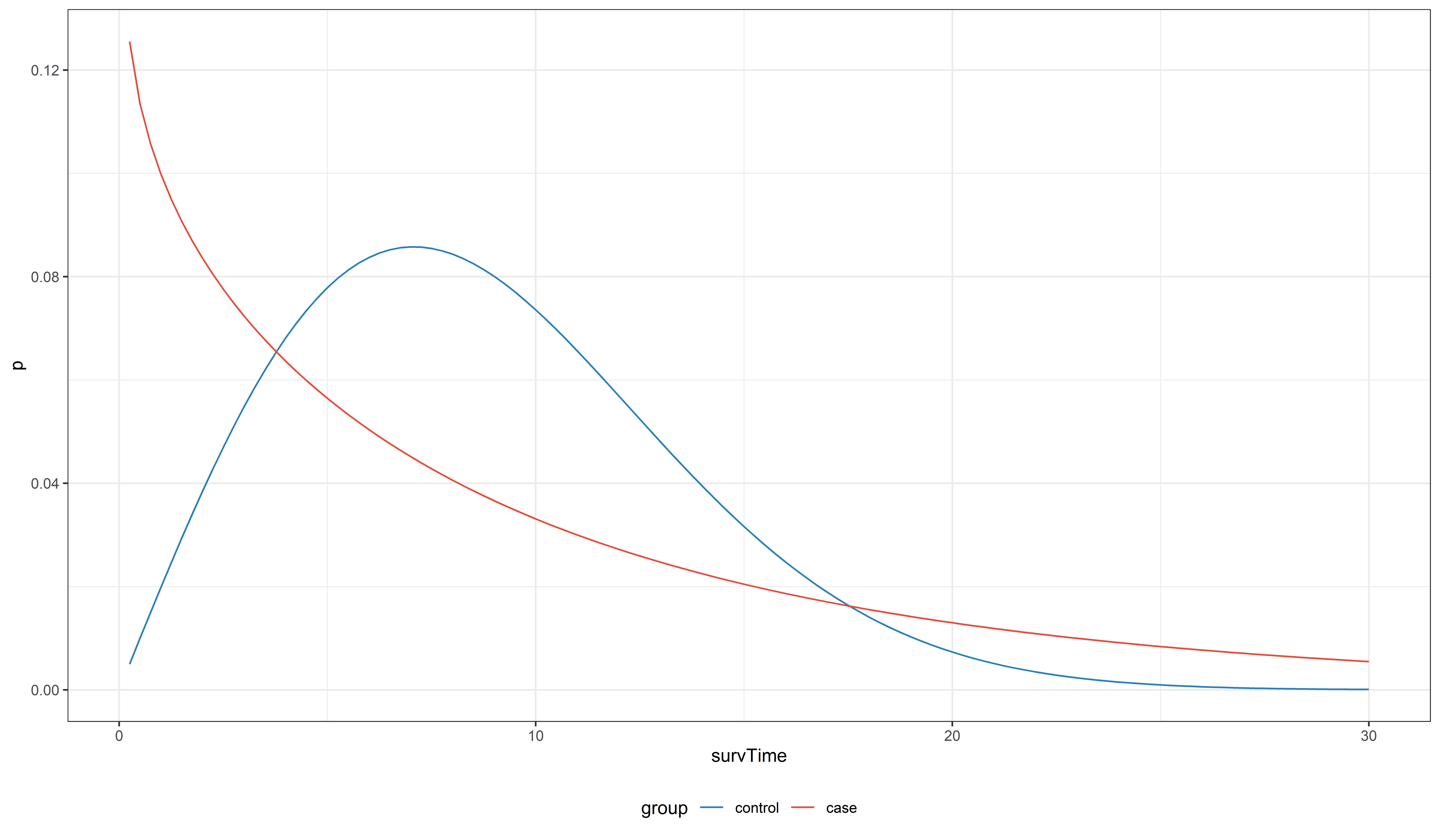
case_scale <- 10So that’s all we need, what we can do now is have a look at what the true distributions look like.
Code
plt_trueSurv <- tibble(survTime = seq(.25, 30, .25)) %>%
mutate(case = dweibull(survTime, case_shape, case_scale),
control = dweibull(survTime, cont_shape, cont_scale)) %>%
gather(case:control,
key = "group",
value = "p") %>%
mutate(group = factor(group, levels = c("control", "case"))) %>%
ggplot(aes(survTime, p, colour = group)) +
geom_path() +
scale_colour_flat()
plt_trueSurvLooks alright, the controls have a distribution that is more to the right while the cases are to the left. Essentially, cases don’t live as long as controls. So now let’s generate some random data
df_model <- tibble(subj = 1:(N * 2),
group = rep(c("control", "case"), each = N),
trueSurv = c(rweibull(N, cont_shape, cont_scale), rweibull(N, case_shape, case_scale))) %>%
mutate(group = factor(group, levels = c("control", "case")))Now we can overlay this onto our true distribution and see what that looks like:
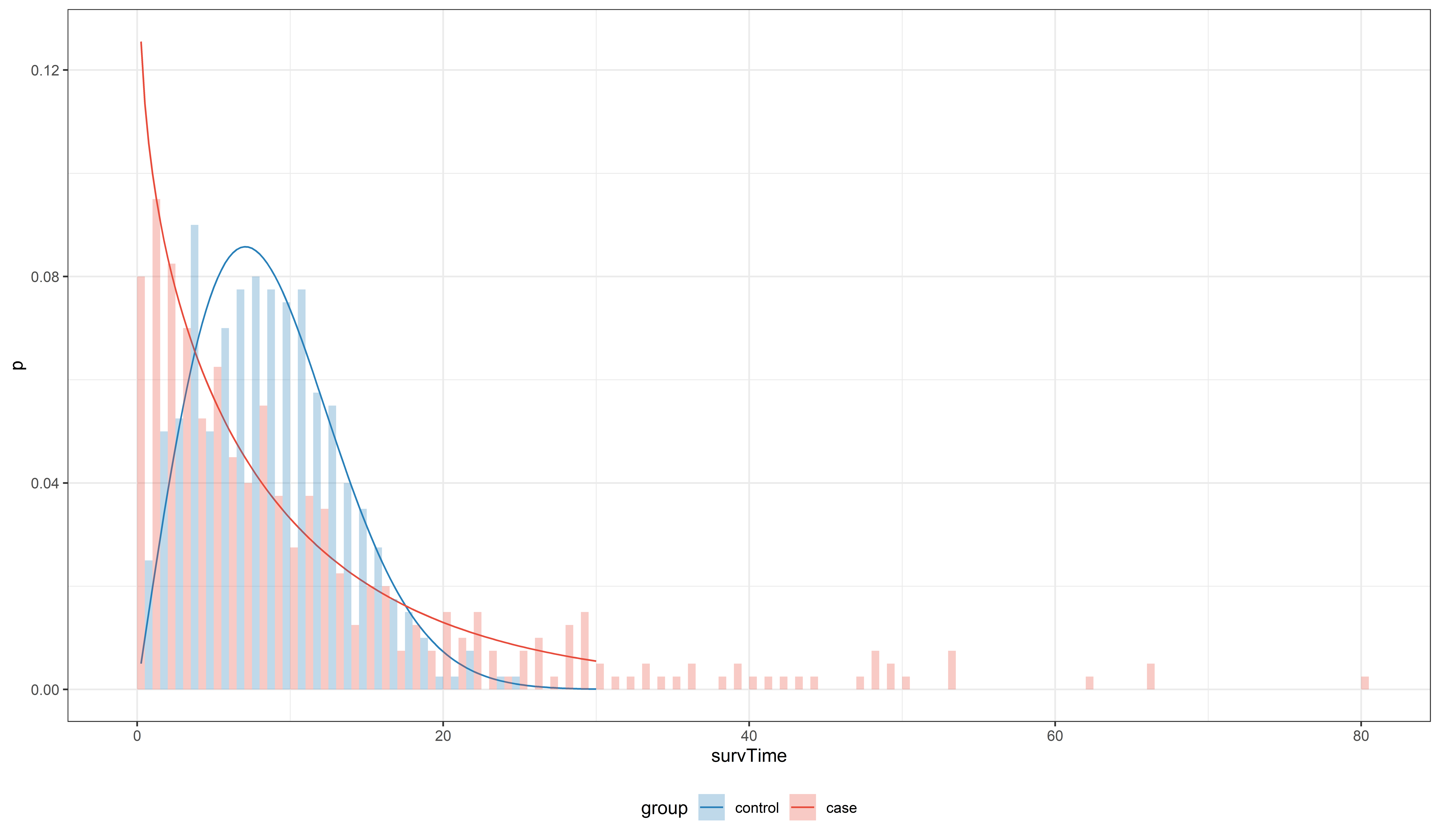
Code
plt_trueSurv_withHist <- plt_trueSurv +
geom_histogram(data = df_model,
aes(x = trueSurv,
y = after_stat(density),
fill = group),
binwidth = 1,
alpha = .3,
position = "dodge",
inherit.aes = F) +
scale_fill_flat()
plt_trueSurv_withHistLooks good enough for our purposes. However, we haven’t exactly added in any censoring just now. So let’s do that, the censoring we’ll add in is called “right-censoring” as we won’t know when the event happened if it happened after the time we were observing for. This is the most common type, and is also the type that has the most work on as far as I can see. So we’ll keep it simple and grounded and just do right censoring.
I’ll do this by randomly sampling between a small number and the true survival time if someone was censored. Each person will have a 50% chance of being censored so it should be roughly even. Though this might be quite a high rate of censoring. We’ll also be assuming that the censoring is uninformative and wouldn’t mean we couldn’t observe the event happening. Unfortunately, no one has found a cure for death, so I think we’re safe to make this assumption, but it is necessary to consider whether a competing event could occur that would stop a subject from being able to experience the event of interest.
df_model <- df_model %>%
mutate(
censTime = runif(nrow(.), .01, .3 * max(trueSurv)),
censored = as.numeric(censTime < trueSurv),
obsTime = ifelse(censored, censTime, trueSurv)
)Keen eyed individuals will notice that I have used 1 to show censoring and 0 to show the event of interest. I know this seems weird, but this is how the package brms expects the censoring information to be presented. I personally also like this, 1 means “this person was censored and so I only know they lived this long event free” and 0 means “this person was not censored and I know when the event happened”.
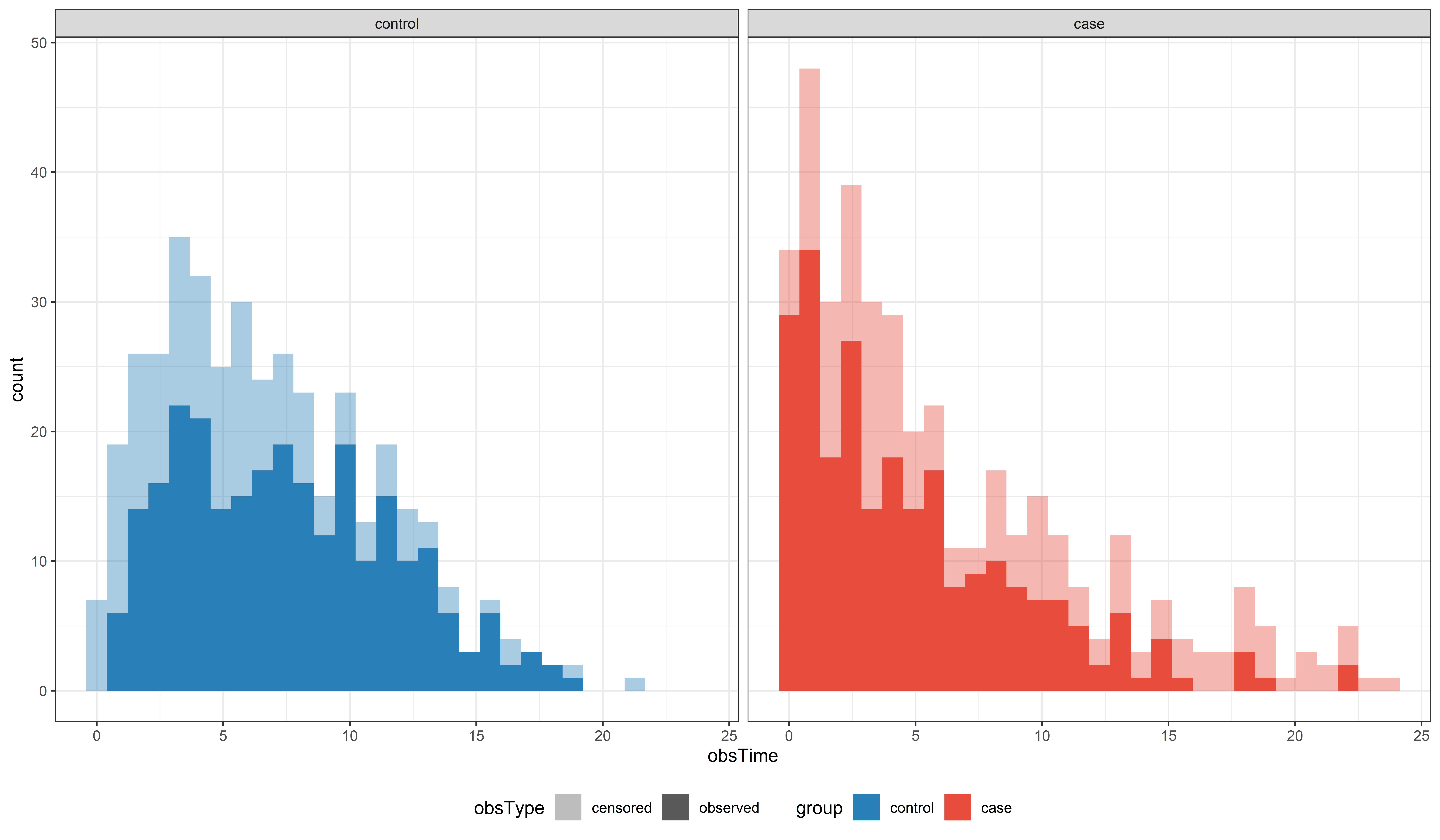
Easy enough, just to confirm this worked, this is what our data look like now:
Code
df_model %>%
mutate(obsType = ifelse(censored, "censored", "observed"),
obsType = factor(obsType, levels= c("censored", "observed"))) %>%
ggplot(aes(obsTime, fill = group, alpha = obsType)) +
geom_histogram() +
scale_fill_flat() +
scale_alpha_manual(values = c(.4, 1)) +
facet_wrap(~group)So, now we have a dataset with a bunch of censoring we can do some analysis and show why we don’t want to ignore censoring.
Doing the analysis
So, there are two ways we will want to analyse these data. The first is a way I would never do, and as such, I’m kind of guessing this is how people would go about doing it. The second is the choice I would make to use, and so I feel more comfortable with this process. Either way, I’ll show where the mistakes creep in and explain why the inference doesn’t make sense.
Ignoring censorship
First, I’ll need to set up the data to be amenable to what I think is the method employed often incorrectly. This method involves looking at who is still alive at some time point and comparing the two groups by way of a logistic regression. In this method, only people that live beyond the time point or who have died by this point are considered in the data. The problem with this is that we are removing a lot of people that could be still alive. We know that they were alive until a certain point, just not until the time point we’re interested in. However, this still gives us information about the likelihood of survival. For example, what if 90% of our population that didn’t die before the time point were known to be alive up until the day before? Hopefully we can recognise that it would be ridiculous to ignore them from our calculations for the probability of survival until time point \(t\)?
So, let’s make the data:
TOI <- 10
df_logi <- df_model %>%
filter(obsTime > TOI | (!censored & obsTime < TOI)) %>%
mutate(logiSurv = obsTime > TOI)The code above simply keeps people that had an observed time that was greater than the Time of Interest (TOI) or those that had experienced the event before the TOI. We then say people were alive if they were in the study longer then the TOI. Sounds simple, but isn’t a great use of data. In this instance we have had to remove 27.25% of our dataset. We also have people that experienced the event almost immediately after the TOI, but we’re ignoring that for this analysis.
Just wanted to highlight that I have many grievances with this approach, if that wasn’t immediately clear.
Now we can fit a simple logistic regression to these data to get the “probability of survival at 5 years”
m_log <- brm(logiSurv ~ group,
family = "bernoulli",
data = df_logi,
chains = 1,
iter = 2000,
warmup = 1000)
SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 1).
Chain 1:
Chain 1: Gradient evaluation took 4.4e-05 seconds
Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.44 seconds.
Chain 1: Adjust your expectations accordingly!
Chain 1:
Chain 1:
Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
Chain 1:
Chain 1: Elapsed Time: 0.137 seconds (Warm-up)
Chain 1: 0.134 seconds (Sampling)
Chain 1: 0.271 seconds (Total)
Chain 1: Now we’ve fit the model, let’s have a look at the output and see what we think
summary(m_log) Family: bernoulli
Links: mu = logit
Formula: logiSurv ~ group
Data: df_logi (Number of observations: 582)
Draws: 1 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 1000
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept -0.65 0.13 -0.91 -0.42 1.00 935 609
groupcase -0.22 0.18 -0.58 0.15 1.00 817 577
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).From this model output, we can see that the likelihood of a case being alive after 10 years is 29.71% [24.59% | 34.95%] compared to the controls who had a likelihood of 34.44% [28.76% | 39.71%]. This might sound reasonable, and reflects that the case group is 0.82 [0.53 | 1.10] as likely to have lived for the observation period. This is all well and good, but we know that this ignores a fair amount of information. So, let’s fit a more appropriate model and see what the estimates look like.
Accounting for censorship
We’ll be fitting a Weibull distribution as we know that this is how the data were generated. In the real world we wouldn’t know this, so we would fit several models and look for different specifications, but that isn’t the point in this document. I’m simply trying to demonstrate the issue with ignoring censoring in these sorts of data.
So, let’s fit the model
m_weib <- brm(bf(obsTime | cens(censored) ~ group,
shape ~ group),
family = "weibull",
data = df_model,
iter = 2000,
warmup = 1000,
chains = 1)
SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 1).
Chain 1:
Chain 1: Gradient evaluation took 0.000622 seconds
Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 6.22 seconds.
Chain 1: Adjust your expectations accordingly!
Chain 1:
Chain 1:
Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
Chain 1:
Chain 1: Elapsed Time: 3.362 seconds (Warm-up)
Chain 1: 3.519 seconds (Sampling)
Chain 1: 6.881 seconds (Total)
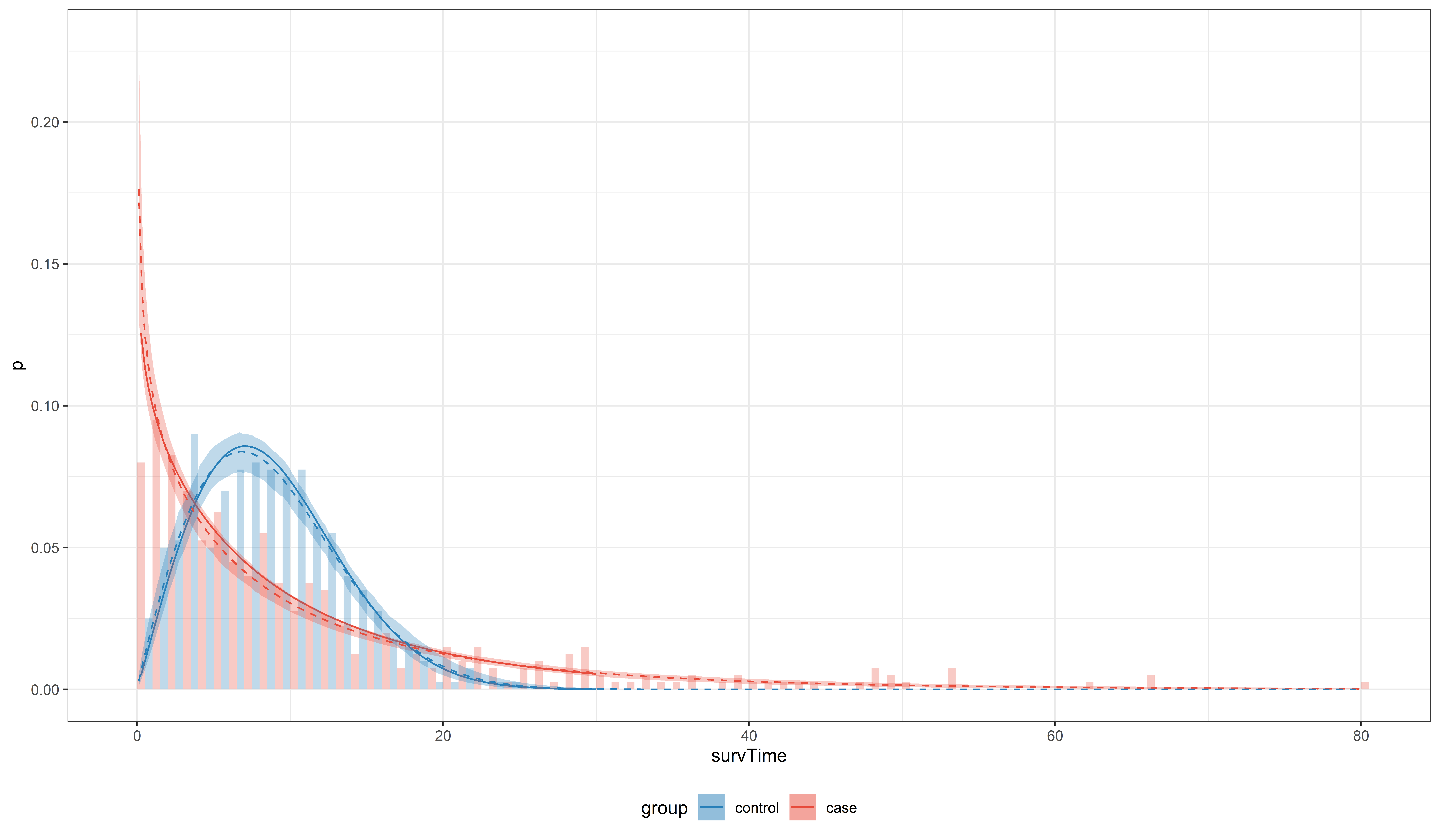
Chain 1: Now we have our new model, we can look at some values in order to compare the estimates. Something that would be interesting first is to compare the model predictions to the real data. We can do this by plotting the estimates for the distribution of survival times over Figure 3 as this is the true distribution of survival times.
Code
# convert from brms parameterisation
get_scale <- function(mu, shape){
exp(mu)/gamma(1 + 1/exp(shape))
}
# get weib post
post_weib <- as_draws_df(m_weib) %>%
select(contains("b_")) %>%
mutate(b_groupcase = b_groupcase + b_Intercept,
b_shape_groupcase = b_shape_Intercept + b_shape_groupcase) %>%
rowid_to_column(var = "iter") %>%
gather(contains("b_"),
key = "Param",
value = "value") %>%
mutate(paramType = ifelse(grepl("shape", Param), "shape", "mu"),
group = ifelse(grepl("groupcase", Param), "case", "control")) %>%
select(-Param) %>%
spread(paramType, value) %>%
mutate(scale = get_scale(mu, shape))
pltData_weib <- post_weib %>%
expand_grid(time = seq(.1, round(max(df_model$trueSurv)), .2)) %>%
mutate(prob = dweibull(time, exp(shape), scale)) %>%
group_by(group, time) %>%
mean_hdci(prob)
plt_trueSurv_withHist +
geom_ribbon(data = pltData_weib,
aes(y = prob, x = time,
ymin = .lower, ymax = .upper,
fill = group),
colour = "transparent",
alpha = .3) +
geom_line(data = pltData_weib,
aes(y = prob, x = time),
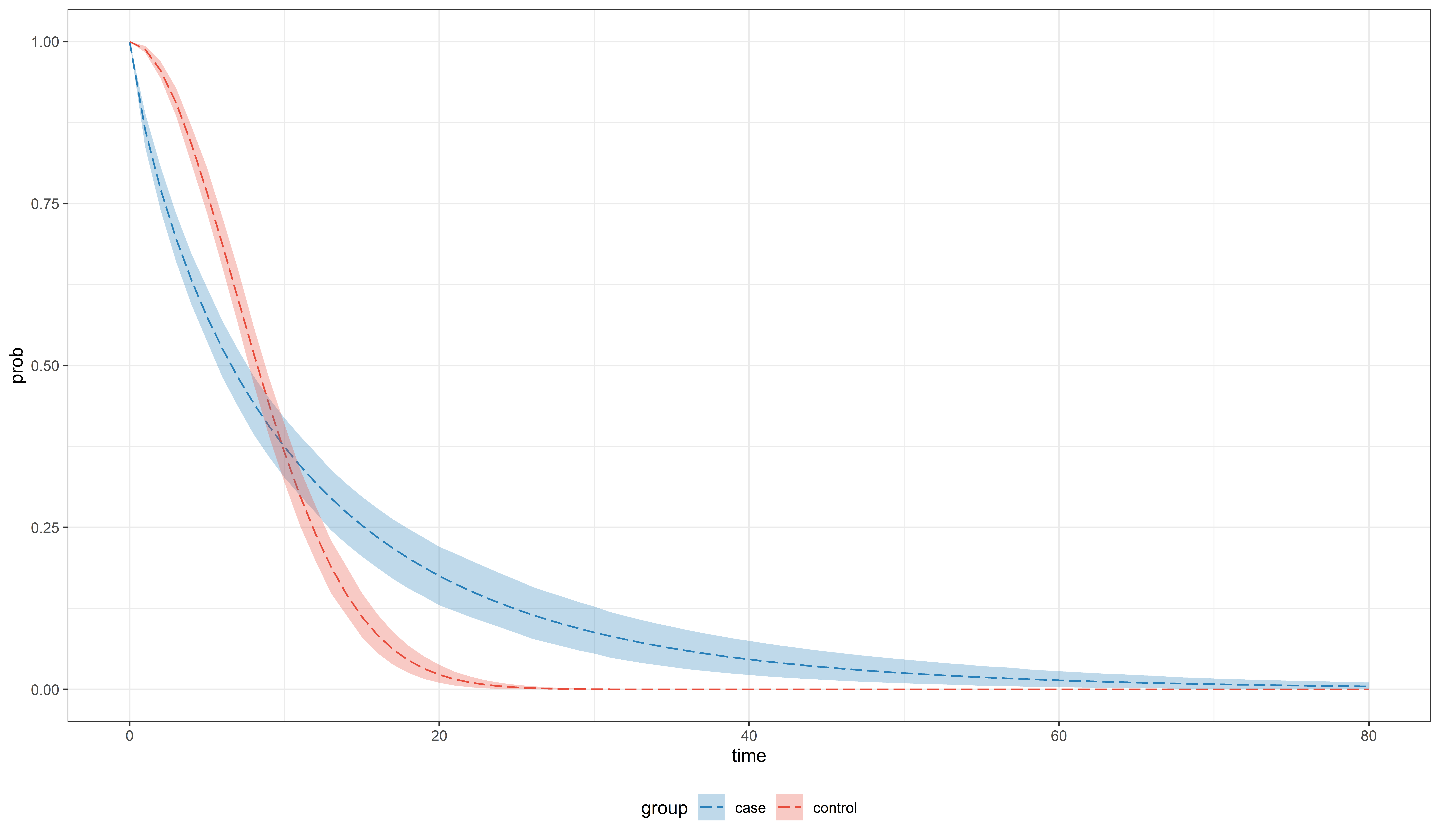
linetype = "dashed")Hopefully we’re fairly convinced that this model has done a pretty good job of retrieving the true values that generated this data, we can now calculate the likelihood that someone would live until 10 years. This is fairly easily done, all we need to do is look a the cumulative density distribution for our specified model and see what the density is at 5 years.
Code
plt_cpdf_weib <- post_weib %>%
expand_grid(time = 0:round(max(df_model$trueSurv))) %>%
mutate(prob = 1 - pweibull(time, exp(shape), scale)) %>%
group_by(group, time) %>%
mean_hdci(prob) %>%
ggplot(aes(time, prob, colour = group, fill = group)) +
geom_path(linetype = "longdash") +
geom_ribbon(aes(ymin = .lower, ymax = .upper),
alpha = .3,
colour = "transparent") +
scale_colour_flat() +
scale_fill_flat()
plt_cpdf_weibFrom this, we can see that the model suggests that the likelihood of survival for cases at 10 years is 37.46% [32.71% | 41.91%] and for controls it is estimated to be 37.46% [32.71% | 41.91%]. This doesn’t appear to match up with the logistic model. The logistic model, although uncertain, suggested that our controls had a lower probability of experiencing the event by 10 years. So which is right?
Since we made this data, we can know everything about it. I’ve waited until now to make this abundantly clear, but we are actually able to show the true survival probability at 10 years by simply calculating this from the initial parameters used to create this data. I’ll leave it as an exercise for you to compare the different approaches in terms of which did a better job of estimating this true value.
writeLines(paste0("control: ", round(exp(-(TOI / cont_scale) ^ cont_shape) * 100, 2), "%\n",
"case: ", round(exp(-(TOI / case_scale) ^ case_shape) * 100, 2), "%"))control: 36.79%
case: 36.79%Unsurprisingly, our model that used the correct distribution makes a better estimate for the true values. So, why did this happen? There was some slight of hand earlier, though I’m sure plenty of people would notice it. When defining the parameters for the simulation, I deliberately picked two shape parameters that would cause the hazard to cross over at some point for the two groups. It isn’t hard to imagine circumstances in which this could happen; for instance, the exposed group (cases) could have a condition that comes with a high risk early on but if managed correctly can result in better outcomes for these people compared to a control group without adequate monitoring. The risk with using a logistic regression for this type of analysis is that the conclusions you make are biased by the time point selected, and the type of censoring present in the data. If we had modelled the survival at 5 years we may have had different conclusions as the ratio of the hazard was not constant over exposure time. In doing a logistic regression on these data, we remove nuance from our understanding of the data itself and could end up making inappropriate conclusions.
A different time point
If we look at Figure 6, we can see that 10 is roughly the point at which the survival functions cross over. So what happens if we look at a different time point, say maybe half this so we have 5 years observation instead? Annoyingly, we’ll have to process the data again for the logistic regression and run the model again in order to get some results, but for the Weibull model we just need to get estimates for a different time point. So, let’s re-run the logistic regression quick.
TOI_2 <- TOI/2
df_logi_2 <- df_model %>%
filter(obsTime > TOI_2 | (!censored & obsTime < TOI_2)) %>%
mutate(logiSurv = obsTime > TOI_2)
m_log_2 <- brm(logiSurv ~ group,
family = "bernoulli",
data = df_logi_2,
chains = 1,
iter = 2000,
warmup = 1000)
SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 1).
Chain 1:
Chain 1: Gradient evaluation took 7.5e-05 seconds
Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.75 seconds.
Chain 1: Adjust your expectations accordingly!
Chain 1:
Chain 1:
Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
Chain 1:
Chain 1: Elapsed Time: 0.147 seconds (Warm-up)
Chain 1: 0.142 seconds (Sampling)
Chain 1: 0.289 seconds (Total)
Chain 1: Now we can simply run what we did before to get the comparisons for the probability of survival until 5 years for both models.
post_m_log_2 <- as_draws_df(m_log_2) %>%
select(contains("b_")) %>%
rowid_to_column(var = "iter")
prob_m_log_2 <- post_m_log_2 %>%
mutate(b_groupcase = b_Intercept + b_groupcase) %>%
gather(contains("b_"),
key = "Param",
value = "logOdds") %>%
mutate(Param = str_remove(Param, "b_"),
prob = plogis(logOdds)) %>%
group_by(Param) %>%
mean_hdci(prob)
paste("control:", reportProb(prob_m_log_2, "Intercept"))[1] "control: 74.04% [69.19% | 78.28%]"paste("case: ", reportProb(prob_m_log_2, "groupcase"))[1] "case: 54.28% [48.94% | 59.40%]"And now the Weibull distribution?
prob_m_weib <- plt_cpdf_weib[["data"]] %>%
filter(time == TOI_2)
paste("control:", reportProb(prob_m_weib, "control", "group"))[1] "control: 76.70% [73.51% | 80.52%]"paste("case: ", reportProb(prob_m_weib, "case", "group"))[1] "case: 57.52% [53.73% | 62.01%]"And now the truth?
paste0("control: ", round(exp(-(TOI_2 / cont_scale) ^ cont_shape) * 100, 2), "%")[1] "control: 77.88%"paste0("case: ", round(exp(-(TOI_2 / case_scale) ^ case_shape) * 100, 2), "%")[1] "case: 58.52%"At this time point, the logistic model isn’t performing too poorly; however, do we really fancy running a model at each time point to estimate the hazard ratio to ensure that we have a constant ratio? I wouldn’t think this would be the case. Further, we have also seen how the estimates for the likelihood of survival can be inaccurate. In the idea of transparency, I believe it is more robust to employ a survival analysis technique that can adequately account for censorship in the data as well as characterise the risk over time and detect non-proportional hazards.
In effect, from this analysis we can see that the choice of time point in the logistic regression can alter the conclusions drawn from the analysis. Having this fixed time point means we run the risk of making incorrect conclusions about the survival rates of different groups due to a almost arbitrary choice about when we decide is the most interesting time point. Having a model that can account for censorship appropriately and also examine survival times is a much more suitable approach.
I can imagine one counter argument being that this approach is too heavy handed if we pick a time point that has very little to no censorship. Realistically, I guess this is a decent argument if you have a strong reason to believe that a particular time point is the most important aspect of the analysis. However, my counter question would be: do you really want to risk being wrong about the time point? And also, isn’t it nice to use a slightly more sophisticated approach that can offer more detailed insights? These are kind of rhetorical questions, but feel free to disagree with me.
Final thoughts
When trying to understand complex questions, we do sometimes have to use complex methods. Censorship in our data isn’t something we want, and in an ideal world there would be no loss to follow up. However, we don’t live in this ideal world and instead have to settle for decent estimates of our measures and incomplete datasets. Additionally, even in an ideal world, looking at survival over time using these methods still offers more insight than a more simple logistic regression does. In my opinion, we should treat the questions we’re asking with respect and attempt to answer them using the most appropriate methods. Settling for “second best” (perhaps not even second best, sometimes just flat out wrong) shouldn’t be the end goal for any analyst. Additionally, isn’t it important to know “when” events happen anyway? For example, we could have all the events in one group happen very early on while the other group had the events happen at random time points between the start and end of follow up?
Anyway, that’s my understanding of survival analysis and the problems with applying logistic regression to censored data. Hopefully that’s been interesting and helpful. A paper that’s worth reading on this subject can be found here. They make a very good case for using survival analysis when approaching problems like this and illustrate why it’s important that we care about this. There’s a good line where they say that in reality, with enough follow up, all survival is 0%. If my work above did nothing to convince you it’s important to care about the timing of events and censorship, then hopefully that one sentence is enough to convince you.
sessionInfo()R version 4.4.2 (2024-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United Kingdom.utf8
[2] LC_CTYPE=English_United Kingdom.utf8
[3] LC_MONETARY=English_United Kingdom.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United Kingdom.utf8
time zone: Europe/London
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] see_0.10.0 brms_2.22.11 Rcpp_1.0.14 tidybayes_3.0.7
[5] lubridate_1.9.4 forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4
[9] purrr_1.0.4 readr_2.1.5 tidyr_1.3.1 tibble_3.2.1
[13] ggplot2_3.5.2 tidyverse_2.0.0
loaded via a namespace (and not attached):
[1] gtable_0.3.6 tensorA_0.36.2.1 QuickJSR_1.7.0
[4] xfun_0.51 htmlwidgets_1.6.4 processx_3.8.6
[7] inline_0.3.21 lattice_0.22-6 callr_3.7.6
[10] tzdb_0.4.0 ps_1.9.0 vctrs_0.6.5
[13] tools_4.4.2 generics_0.1.3 stats4_4.4.2
[16] parallel_4.4.2 pkgconfig_2.0.3 Matrix_1.7-1
[19] checkmate_2.3.2 distributional_0.5.0 RcppParallel_5.1.10
[22] lifecycle_1.0.4 farver_2.1.2 compiler_4.4.2
[25] Brobdingnag_1.2-9 munsell_0.5.1 codetools_0.2-20
[28] htmltools_0.5.8.1 bayesplot_1.12.0 yaml_2.3.10
[31] pillar_1.10.2 arrayhelpers_1.1-0 StanHeaders_2.32.10
[34] bridgesampling_1.1-2 abind_1.4-8 nlme_3.1-166
[37] rstan_2.32.7 posterior_1.6.1 tidyselect_1.2.1
[40] digest_0.6.37 svUnit_1.0.6 mvtnorm_1.3-3
[43] stringi_1.8.7 reshape2_1.4.4 labeling_0.4.3
[46] fastmap_1.2.0 grid_4.4.2 colorspace_2.1-1
[49] cli_3.6.4 magrittr_2.0.3 loo_2.8.0.9000
[52] pkgbuild_1.4.7 withr_3.0.2 scales_1.3.0
[55] backports_1.5.0 timechange_0.3.0 rmarkdown_2.29
[58] matrixStats_1.5.0 gridExtra_2.3 hms_1.1.3
[61] coda_0.19-4.1 evaluate_1.0.3 knitr_1.49
[64] ggdist_3.3.2 rstantools_2.4.0.9000 rlang_1.1.5
[67] glue_1.8.0 rstudioapi_0.17.1 jsonlite_1.9.0
[70] plyr_1.8.9 R6_2.6.1